In Silico Co-evolution of Transcription Factor Binding Domans and Cognate Binding Sites
Computational modeling of TF/BS co-evolution allows us to test hypotheses about the design principles underlying transcriptional regulation.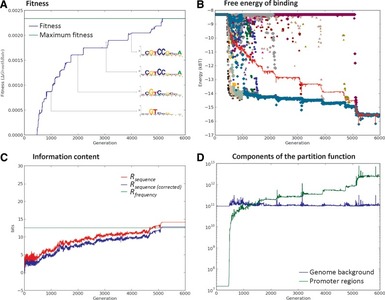
Computational modeling of TF/BS co-evolution allows us to test hypotheses about the design principles underlying transcriptional regulation.
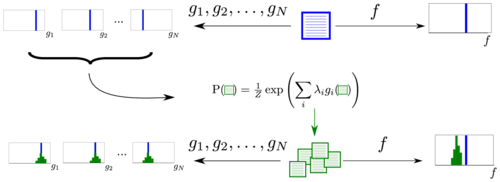
MaxEnt bootstrapping provides a general framework for assessing the significance of arbitrary statistics of sequence motifs.