In Silico Co-evolution of Transcription Factor Binding Domans and Cognate Binding Sites
The evolution of simple transcriptional regulatory networks has been studied before by means of explicit simulation of TFs and their binding sites, leading to fundamental insights into the informational constraints faced by such networks. In this work we generalize this approach by developing an evolutionary simulator that incorporates biophysical modeling and addresses several caveats of previous attempts.
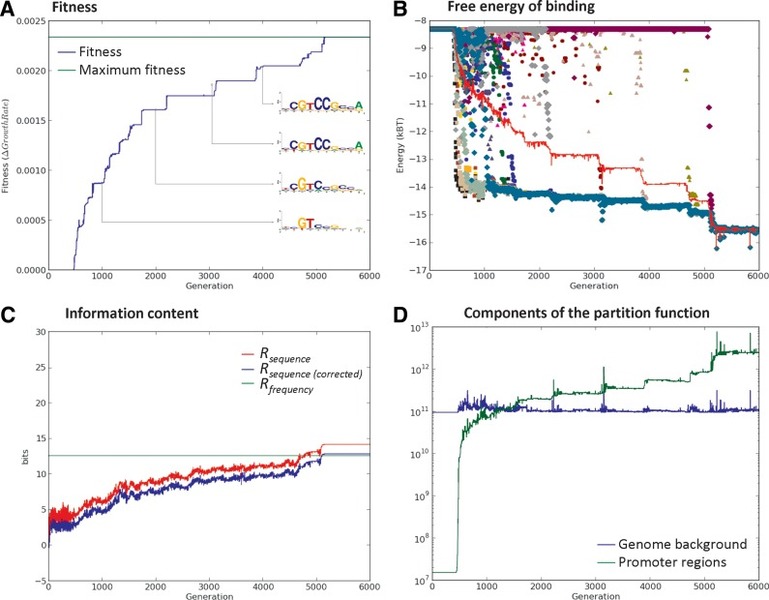
The Evolutionary Simulator of Transcriptional Regulatory Motifs (ESTReMo) uses a genetic algorithm backbone to simulate the evolution of transcriptional regulatory networks using a modular approach. In ESTReMo, each organism contains two evolvable components: a TF model and a set of promoter regions for target genes. The TF is modeled as a feed-forward artificial neural network that operates on the set of target promoter regions and on a fixed genomic background. In each generation, the TF model is used to compute the free energy of binding for all positions in target promoter regions and in a randomly sampled subset of the genomic background. The occupancy of each promoter position is computed according to a statistical mechanical distribution on an effective background generated by scaling up the randomly drawn samples. These occupancies are then used to determine the expression level for each target gene following a given expression model. Finally, organism fitness is evaluated as the sum of cost-benefit differences for each target gene and its desired expression level using an empirical fitness model.
ESTReMo can be used to analyze the impact of differential regulatory demands on the information content of TF-binding motifs by means of evolutionary simulations. Our results show a logarithmic dependence of the evolved information content on the occupancy of target sites and indicate that TFs may actively exploit pseudo-sites to modulate their occupancy of target sites. In regulatory networks with differentially regulated targets, we observe that information content in TF-binding motifs is dictated primarily by the fraction of total probability mass that the TF assigns to its target sites, and we provide a predictive index to estimate the amount of information associated with arbitrarily complex regulatory systems. We observe that complex regulatory patterns can exert additional demands on evolved information content, but, given a total occupancy for target sites, we do not find conclusive evidence that this effect is because of the range of required binding affinities.
