Page Not Found
Page not found. Your pixels are in another canvas.
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Page not found. Your pixels are in another canvas.
About me
This is a page not in th emain menu
One of the most influential lectures I ever caught in grad school was Ryan Gutenkunst’s talk on sloppy models. Model sloppiness, first developed by Jim Sethna and his students and collaborators, is a ubiquitous phenomenon in mathematical models for the natural sciences. Often, especially in biology, a model of a system of interest will have many more parameters to be fit than observations to fit them with: a scenario like eighty parameters and six data points is not uncommon. Standard theory tells you that you are doomed here. But if you ignore standard theory and plow ahead, you typically find that the (under)fit model will predict future experiments perfectly well. How could that possibly work?
Shared ownership is supposed to align incentives. If two founders each own 50% of a given company, then they will tend to want the same things for that company. But consider a founder who owns 99% of one company and an investor who owns 1% of 99 companies– do these actors always have the same incentives? Not necessarily. To see why, consider the following model.
Next week I’ll be giving a guest lecture at MIT for 15.S08, Advanced Data Analytics and Machine Learning in Finance, sketching an overview of modern speech-to-text systems and offering some lessons learned from building Scribe.
Blog coming soon.
Published in Journal 1, 2009
This paper is about the number 1. The number 2 is left for future work.
Recommended citation: Your Name, You. (2009). "Paper Title Number 1." Journal 1. 1(1). http://academicpages.github.io/files/paper1.pdf
Published in Journal 1, 2010
This paper is about the number 2. The number 3 is left for future work.
Recommended citation: Your Name, You. (2010). "Paper Title Number 2." Journal 1. 1(2). http://academicpages.github.io/files/paper2.pdf
Published in Journal 1, 2015
This paper is about the number 3. The number 4 is left for future work.
Recommended citation: Your Name, You. (2015). "Paper Title Number 3." Journal 1. 1(3). http://academicpages.github.io/files/paper3.pdf
Computational modeling of TF/BS co-evolution allows us to test hypotheses about the design principles underlying transcriptional regulation.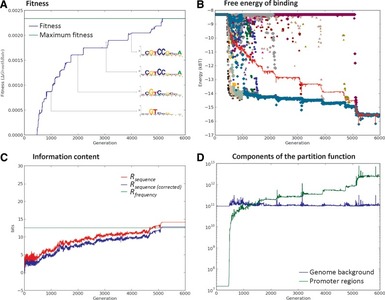
MaxEnt bootstrapping provides a general framework for assessing the significance of arbitrary statistics of sequence motifs.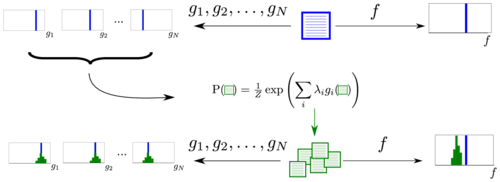
Presented research to the general public and advised middle school children on science fair projects.
Guest lecture at the Maryland Institute College of Art on evolutionary game theory, focusing on the rock-paper-scissors game in well-mixed and spatially resolved populations.
Guest lecture on category theory for high school students.
An introductory talk on the aims and methods of computational biology for a high school audience.
In grad school I was thrice invited by the UMBC Biology Council of Majors to debate various bioethical topics. Although I was technically undefeated (as measured by audience polling), the truth is of course not determined by a vote. I thank BIOCOM and all of my opponents for the occasions to explore and defend my views.
Talk and live coding demonstration on elementary gene finding algorithms.
Guest talk to The Artemis Project on computational biology.
Eight week introduction to programming with Python.
Six week introductory course on Python and elementary bioinformatics using Project Rosalind.
Joint with Towson University Applied Math Laboratory.
Joint with Sara Miller and Dennis Howell.
Transcriptional regulation is the most ubiquitous mechanism for control of gene expression, motivating the development of quantitative models for the kinetics and evolutionary dynamics of transcription factors. Occupancy probabilities for transcription factor binding sites are Fermi- Dirac distributed; while it is commonly granted that the chemical potential of such a distribution is well-modeled by an ideal gas approximation, it is not difficult to solve for it numerically. We do so for 27 transcription factors in Escherichia coli and show that in many cases the ideal gas approximation can diverge sharply from the exact solution, leading to underestimation of site occupancies by orders of magnitude. We conclude that researchers may observe qualitatively different behaviors in models of transcriptional regulation which admit approximate versus exact chemical potentials.
Guest lecture to 15.S08, Advanced Data Analytics and Machine Learning in Finance.
TA for Undergraduate Course, UMBC, 2011
Served as teaching assistant for upper level cell biology course, Ran discussion sections, designed quizzes, and supervised undergraduate teaching assistants.
TA for Undergraduate Course, UMBC, 2012
Served as teaching assistant for senior bioinformatics seminar. Graded, held office hours, ran labs and guest lectured.
TA for Undergraduate Course, UMBC, 2012
Served as teaching assistant for upper level bioinformatics course. Graded, held office hours and guest lectured.